July 17, 2020
OntoloBridge - A Novel Platform to Connect Users and Developers of Scientific Vocabularies
Now Available via the BioPortal Ontology Repository
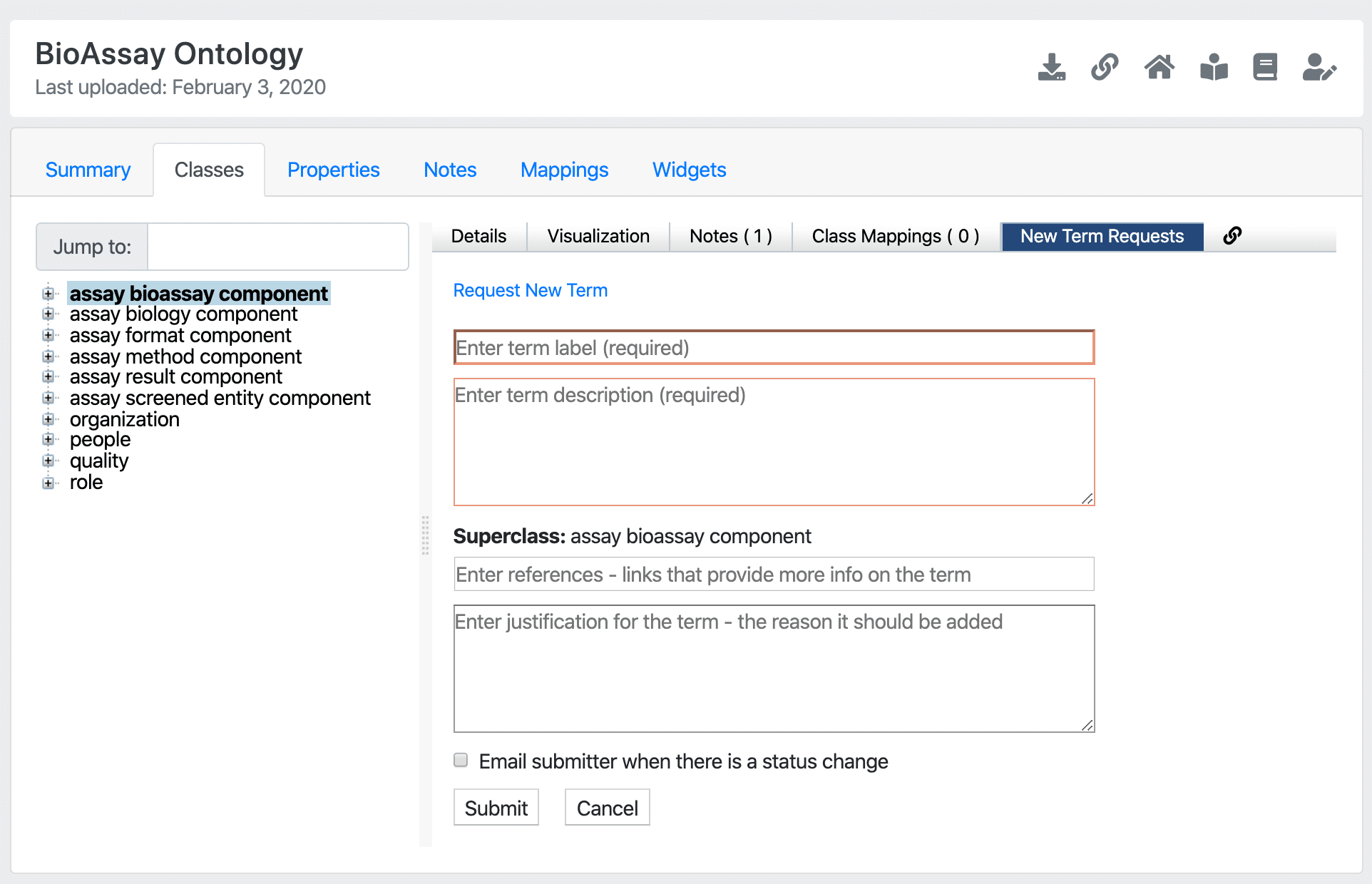
Screenshot of OntoloBridge in the BioPortal
OntoloBridge is now available on BioPortal! The project, funded by a collaborative U01 grant between Stanford University, University of Miami, and Collaborative Drug Discovery, was designed to bridge the gap between regular users of controlled scientific vocabularies and the creators of underlying ontologies.
Ontologies are descriptions of scientific terms and their relationships with human readable and formal (logical) definitions. Ontologies thus serve as smart thesauri that define and interrelate scientific terms. In the age of Big Data, formal vocabularies are more important than ever to enable find, access, integrate and reuse datasets and other digital research objects, in other words to make data FAIR (findable, accessible, interoperable, reusable).
OntoloBridge is a new technology that enables a more convenient and semi-automated process for ontology developers to update and expand their ontologies with curated new terms while accepting community suggestions.
Encouraging and handling community input is a crucial feature in widespread adoption of ontologies. Scientists should not have to choose between selecting less favorable or unsuitable vocabularies or avoiding semantic annotations of their data all together. OntoloBridge allows researchers to connect directly with ontology owners to request new terms that would benefit both their immediate workflow and the scientific community at large.
Initial user reaction has been profoundly positive. Feedback generated from using Collaborative Drug Discovery's bioassay annotation tool, BioAssay Express (www.bioassayexpress.com), in conjunction with the OntoloBridge integration has led to increased productivity by streamlining semantic annotation of bioassays. Users can suggest ontology terms on the fly, initiating a feedback system between curators and ontology experts.
OntoloBridge currently serves the BioAssay Ontology (BAO), Drug Target Ontology (DTO), Protein Ontology (PR), Cell Line Ontology (CLO), and the Coronavirus Infectious Disease Ontology (CIDO). Our team is in the process of expanding the reach of the platform to additional ontologies. If you want your ontology to work with OntoloBridge, please contact info@ontolobridge.org. (If you want your ontology added to BioPortal, contact jgraybeal@stanford.edu)
About this grant
The U01 grant supports collaborative substantial programmatic development between NIH and awarded institutes. Award Number U01LM012630 from the National Library of Medicine as described on NIHReporter supports this project. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. More information at: https://projectreporter.nih.gov/project_info_description.cfm?aid=9747967&icde=50608877.
About CDD
CDD’s (https://www.collaborativedrug.com/) flagship product, CDD Vault®, is a modern web application for your chemical registration, assay data management and SAR analysis. CDD Vault® is a hosted database solution for secure management and sharing of biological and chemical data. It lets you intuitively organize chemical structures and biological study data, and collaborate with internal or external partners through an easy to use web interface. CDDVault® is continuously growing its technology most recently to include ELN, Model and API functionality. A complete list of the 58 CDD publications/patents can be found online on our resources page: https://www.collaborativedrug.com/pages/resources.
For more information contact:
Collaborative Drug Discovery, (650) 204-3084, info@collaborativedrug.com.
About Schürer Lab at the University of Miami
The Schürer lab operates at the Department of Pharmacology in the Miller School of Medicine (http://pharmacology.med.miami.edu/) and the Institute for Data Science and Computing (iDSC) (https://ccs.miami.edu/focus-area/drug-discovery/) at the University of Miami. The primary research theme at the Schürer group is systems drug discovery. We integrate and model small molecule-protein interaction, systems biology ‘omics,’ and chemistry data to improve translation of disease models into novel functional small molecules. Using distributed and parallelized big data analytics, bio- and chemoinformatics tools we build sophisticated modeling pipelines to understand and predict drug mechanism of action, promiscuity and polypharmacology with a particular focus on kinases and epigenetic bromodomain reader proteins with application to cancer and other diseases. In several focused as well as larger-scale projects, we develop formal ontologies (e.g. BioAssay Ontology, Drug Target Ontology), data standards, and end-user multi-tier software applications. We have several drug discovery collaborations ranging from cancer to neurological disorders.
To physically make and test the most promising small molecules, we are developing computationally-optimized synthetic routes and we use parallel synthesis technologies to make small compound libraries. The combination of computational and chemistry methodologies accelerated lead optimization and the development towards clinical compounds.
The group participates in three national Consortia, the Library of Integrated Network-based Cellular Signatures (LINCS) project (https://lincsproject.org/), the Big Data to Knowledge (BD2K) program (https://datascience.nih.gov/bd2k), and the Illuminating the Druggable Genome (IDG) project (https://druggablegenome.net/). Most recently we are also part of the recently established NIH Data Commons.
Schürer publications: https://goo.gl/TsNof9
Contact: Stephan Schürer, PhD, University of Miami, +1-305-243-6552, sschurer@miami.edu.
About Stanford University’s Center for Biomedical Informatics Research
The Stanford Center for Biomedical Informatics Research (BMIR; http://bmir.stanford.edu/) studies the development and evaluation of advanced computational methods to enhance biomedicine. BMIR is home to the Center for Expanded Data Annotation and Retrieval (CEDAR), which offers modular, REST-accessible software to accelerate the specification, collection, and publication of scientific metadata, while increasing the quality, quantity, and interoperability of the resulting metadata descriptions. From ontology creation (Protégé), to ontology and vocabulary dissemination (BioPortal), to semantic metadata management (CEDAR Workbench), BMIR provides transformative open source products and services used by millions of researchers.
References to all our projects’ publications are provided at https://metrics.stanford.edu/people/mark-musen.
Media Contact: Mark Musen, MD, PhD, Stanford University, +1-650-725-3384, musen@stanford.edu.
Other posts you might be interested in
View All Posts
CDD Blog
6 min
February 14, 2018
Public-Private Partnership to Facilitate Ontology Use and Application
Read More
CDD Blog
3 min
June 17, 2015
CDD Awarded $1.5 Million Grant to Simplify Encoding of Bioassays with Automated Annotation Technology
Read More
CDD Blog
3 min
September 2, 2014
CDD Innovation: A New Tool for Annotating Bioassays
Read More


