October 16, 2023
CDD Vault Update (October 2023): Keyword Searching, A New Molecule Pop-Up, ELN Summary-Only Export, Structure Searches Return Mixtures, and New API for Handling Files
Keyword Searching
When using the Keywords search feature in the main Explore Data > Search tab, there is a new “exact” search option available when you select a specific, non-numeric Molecule or Batch field.
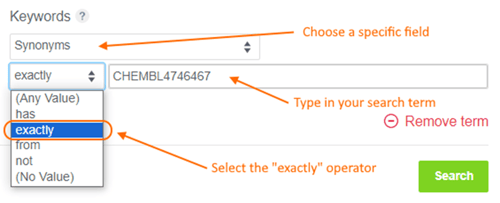
Helpful hint: the algorithm used when selecting the “has” operator was also updated to no longer wrap the search term in quotes, so your search terms are more likely to return a desirable list of matches. This is especially relevant for IDs with multiple special characters and spaces.
Updated Molecule Pop-Up
When clicking on a structure box, the resulting pop-up dialog box has been revamped
- new Size options are available for downloading SVG and PNG files
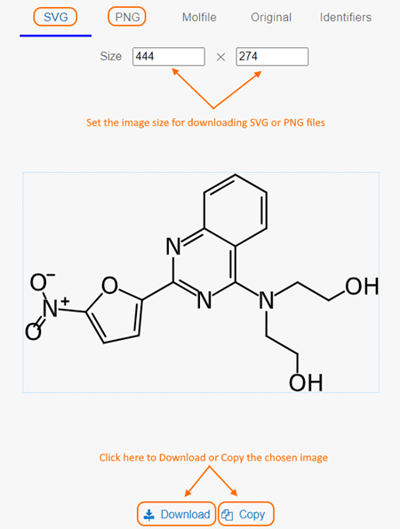
- Molfile and Original options display the file format, with options to Download or Copy

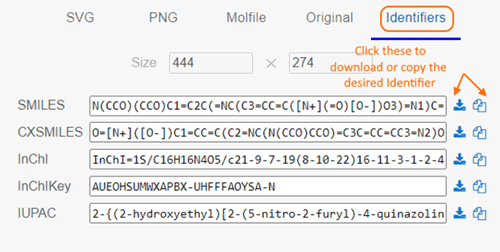
- A new Identifiers section was added with options to Download or Copy these formats:
- SMILES
- CXSMILES
- InChI
- InChIKey
- IUPAC
ELN Summary-Only Export
Vault Administrators have always been able to export a set of ELN Entries from the main ELN Index page. A new “Export summary” option has been added to allow Vault Administrators to export a single summary file.
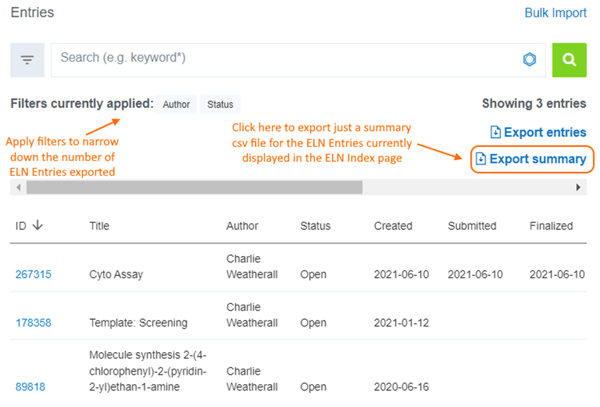
Helpful Hints:
- All system ELN metadata fields plus all user-defined ELN Fields are included in the csv Summary file.
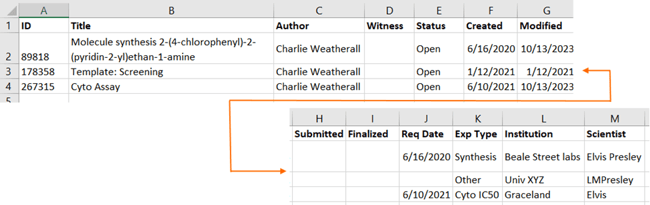
- A new only_summary parameter was added to the GET eln/entries API call to return only this csv Summary file. Simply submit JSON similar to:
{
"author":"6021",
"status":"open",
"only_summary":true,
"async":true
}
Structure Searches Return Mixtures
Structure-based queries now return Mixtures which contain components that match the query.

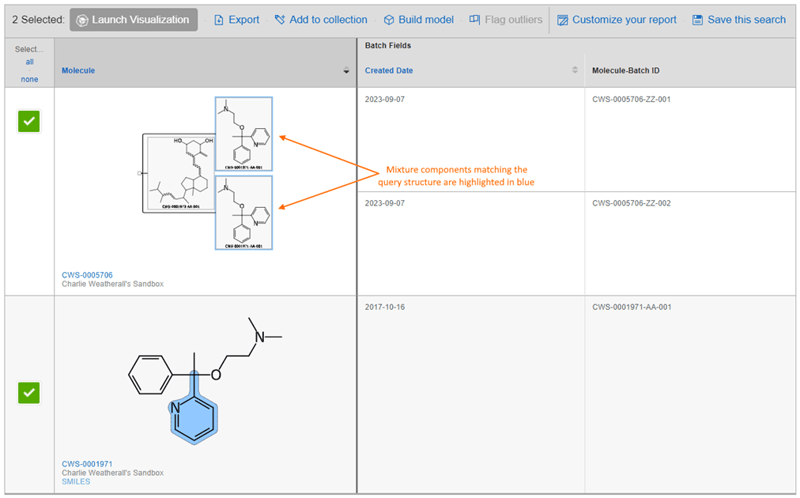
Helpful hints:
- The mixture components must be registered entities in order to be indexed.
- Sequence based searches will also return matching mixtures.
New API for Handling Files
There is a 2-step API workflow now available to programmatically attach files to Molecule/Batch fields and Protocol readouts.
The first step involves using thePOST FilesAPI endpoint to send a file to the CDD Vault server. This step returns an internal file id that must be used in a 2nd API call to attach this file to the desired field/readout.
Step 1
The 1st API call used is POST /api/v1/vaults/<vault_id>/files which returns JSON containing the internal file id.
{
"id": 20167608,
"name": "FlowCyto.png",
"expires_at": "2023-10-13"
}
Helpful Hint: The JSON includes an “expires_at” date indicating when the file will be removed from the CDD Vault server if it’s not subsequently attached to a field/readout or object.
Step 2
The next API call attaches the file to the desired field/readout. This step uses the Molecules, Batches or Readout_Rows endpoints.
As an example, if the file just POSTed needs to be attached to an existing Batch field, use this API call:
PUT https://app.collaborativedrug.com/api/v1/vaults/<vault_id>/batches/<batch_id>
And pass JSON like this:
{"batch_fields":
{"DataFile": 20167608}
}
As another example, if the file just POSTed needs to be attached to an existing Protocol readout row, use this API call:
PUT https://app.collaborativedrug.com/api/v1/vaults/<vault_id>/readout_rows/<readout_row_id>
And pass JSON like this:
{"readouts":
{"1146750": {"value": "20167608"}}
}
This blog is authored by members of the CDD Vault community. CDD Vault is a hosted drug discovery informatics platform that securely manages both private and external biological and chemical data. It provides core functionality including chemical registration, data visualization, inventory, and electronic lab notebook capabilities.
Other posts you might be interested in
View All Posts
CDD Blog
3 min
April 14, 2025
Let’s Talk Security - Why a Bug Bounty May Be More Valuable Than a Penetration Test
Read More
CDD Vault Updates
7 min
April 10, 2025
CDD Vault Update (April 2025): Biphasic Curve Fit, Import Parser Sections, Custom Calculation Functions, Generate Inventory Labels, Inventory Admin Permission
Read More
CDD Blog
9 min
April 8, 2025
Drug Discovery Industry Roundup with Barry Bunin — April 8th, 2025
Read More


