July 28, 2023
CDD Vault Update (July 2023): Annotate Assays using Protocol Forms, Filter and Compare Protocols, and Define Mixture Uniqueness
Using Protocol Forms to Annotate Assays/Runs
Assay analytics plays a vital role in an organization's ability to understand the scientific domain covered by your current assays, identify potential gaps, and develop new/novel assays to advance your research. CDD Vault is an essential platform for managing your assay analytics workflow.
Biological assay data is managed in CDD Vault using Protocol “containers” that usually have many Runs (think experiments) associated with them. These Protocol definitions (created by you) have Readouts that specify the type and location of data points for a given assay. Protocol definitions can also contain custom calculations and graphs derived from these Readouts.
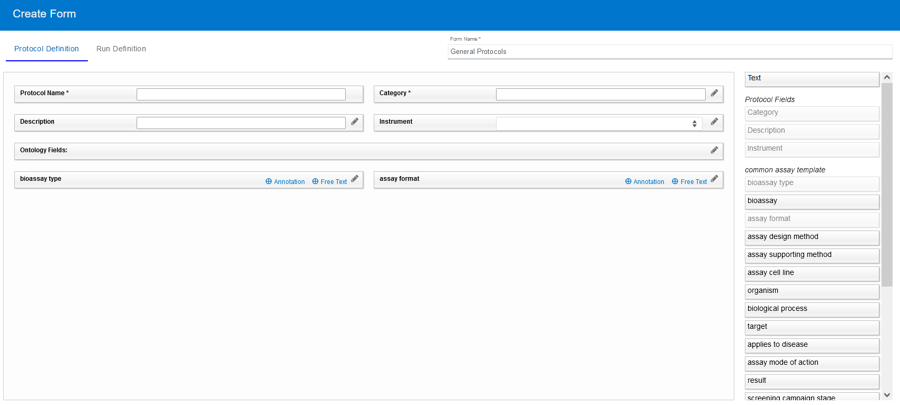
To fully annotate your assays, Vault Administrators may create Protocol- and Run-level fields which are available when defining a new Protocol and Run. Taking annotation a leap forward, Protocol Forms now streamlines the creation of precisely structured Protocols and Runs by offering users a custom set of predefined fields and/or ontology terms that are relevant for each specific type of assay.
The Common Assay Template is a set of ontology terms provided by default for all users. This includes common assay descriptors which can be used to better annotate your assays (FAIR). Forms allow you to pick and choose which fields are relevant for each category of assay. You can set business rules, provide default values, and even lock values for each field thus providing more detail about that type of assay without creating more work for the person defining the Protocol.
Once your Vault Administrator creates Forms, they can be selected when creating new Protocol definitions and subsequent Runs.
The implementation of Protocol and Run Forms within your assay analytics strategy can:
- Bring assay awareness to each scientist so key details are captured
- Ensure assays are fully and consistently annotated so data isn’t lost over time
- Shorten Protocol creation time by providing specific fields instead of generic text boxes
- Elucidate what assays have been performed and how they relate to each other, which can be key for QSAR/AI models
- Share assay details with collaborators, CROs, publications, etc. using a common universal vocabulary
- Make assay data precisely searchable: find protocols with specific characteristics, either using specific terms or branch categories
Helpful Hints:
- Create one form for each type of assay. Form names should be short and descriptive so they’re easy to navigate when defining a new protocol, e.g. Enzyme, Cell, Animal, Tox.
- Protocol Forms are made up of two definitions, Protocol and Run. Ontology terms may be used in either definition while fields are specific to their definition.
- When defining your fields, consider whether a value will change over time (across Runs) and whether the value is used in data processing:
- Protocol fields - describe the assay and do not change over time
- Run fields - may change over time and includes environment variables that do not directly impact the data processing
- Condition-based Readout Definitions - variables that are used in the calculation and processing of data
- Alternate ontologies can be used and multiple templates can be stored within CDD Vault and used in different forms. Contact CDD support to import your custom ontologies.
- Forms can be modified over time. Since Forms only impact the display of fields, users can apply different Forms to existing Protocol definitions.
Filter and Compare Protocol Definitions
An advanced search option has been added to the Protocol index page. Filter by protocol fields and ontology terms, and view Protocols in a property grid.
The new filter layers can be used when more specificity is needed beyond a simple keyword search. Click the “Advanced search…” link and “Add a new layer”.
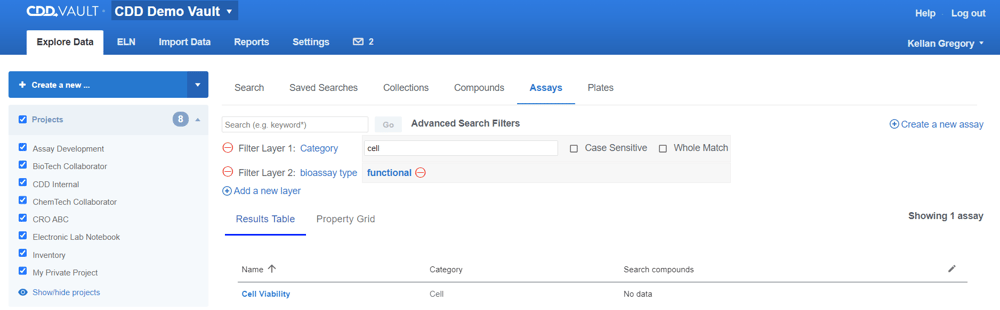
Protocol fields are listed at the top of the filters list, followed by ontology terms. The gray background indicates when a field has a value based on the available Protocols.
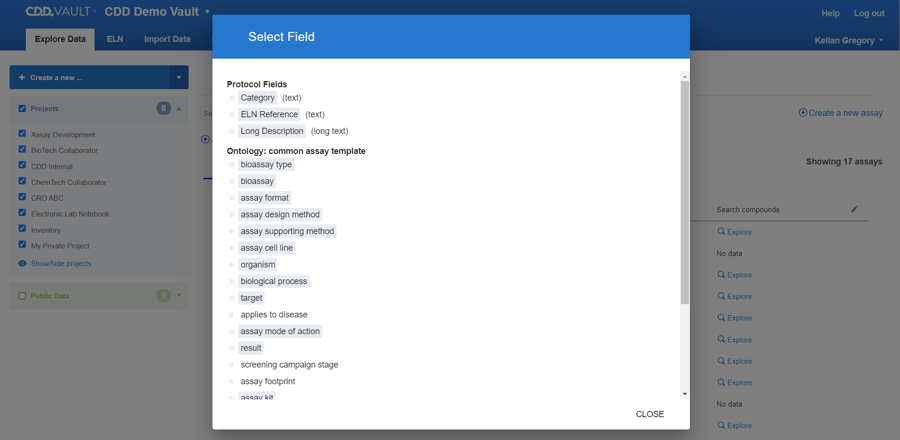
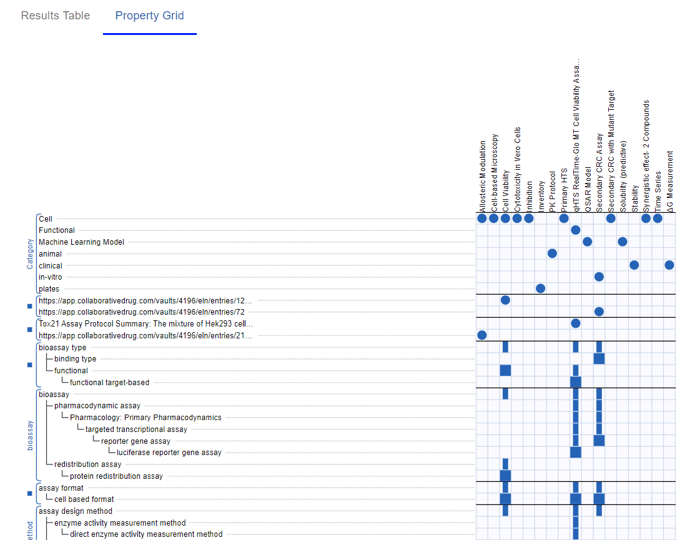
You can specify keywords, number ranges, and ontology terms. The Table updates in real time and you have the option to view your Protocols in a Grid view.
Defining Uniqueness for Mixtures
Accurately representing Mixtures has historically been difficult. Mixtures are often present in boutique combination drugs, cosmetic formulations, complex food additives, and natural product pesticides. As such, they are an important part of numerous scientific domains and the ability to represent Mixtures in an informatics platform is advantageous.
Defining uniqueness across Mixtures involves characterizing the specific combination of components. Each organization has different requirements when it comes to uniqueness so records are created and maintained in the desired hierarchy, and the results can be easily digested. It also facilitates the identification of novel Mixtures with distinct properties or therapeutic potential and the effect of singular component changes on such properties.
CDD Vault now provides a configurable registration system for Mixtures which Vault Administrators can control. In the Settings > Vault > General tab, a Vault Administrator can set the "Mixture equivalence" to: None, Component name (text), Molecule, Molecule Batch:
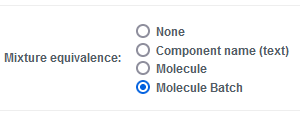
None
When the “None” option is selected, CDD Vault does not perform any uniqueness checking on Mixtures and duplicate Mixtures are allowed.
Component name (text)
When the “Component name” option is selected, CDD Vault will define unique Mixtures as those having the same set of Components based on the Component Name text values. A new Mixture being registered with the exact same Component Names will be added as a new Batch of the existing Mixture.
Molecule
When the “Molecule” option is selected, CDD Vault will define unique Mixtures as those having the same combination of components based on registered Molecules. The records may or may not contain chemical structures or sequences. As long as the registered Molecules are the same, the batch and order do not matter.
Molecule Batch
When the “Molecule Batch” option is selected, CDD Vault will define unique Mixtures as those having the same combination of components with identical Molecule-Batch IDs. The specific Batches of each component will determine whether a new Mixture record is created.
Helpful Hint: If your Mixture contains components which lack the specificity of the given Mixture Equivalence, the next tier down is used. This way you can still have a Mixture where the active component is a registered Molecule but the other components are common reagents which are not registered.
This blog is authored by members of the CDD Vault community. CDD Vault is a hosted drug discovery informatics platform that securely manages both private and external biological and chemical data. It provides core functionality including chemical registration, data visualization, inventory, and electronic lab notebook capabilities.
Other posts you might be interested in
View All Posts
CDD Vault Updates
10 min
April 18, 2025
CDD Vault Update (April 2025 #2): Pharmacokinetic (PK) and Michaelis-Menten Kinetics (Km/Kd) Curve Fit Equations, Donut Charts, TIFF Image Previews, and Parallel Reactions
Read More
CDD Blog
3 min
April 14, 2025
Let’s Talk Security - Why a Bug Bounty May Be More Valuable Than a Penetration Test
Read More
CDD Vault Updates
7 min
April 10, 2025
CDD Vault Update (April 2025): Biphasic Curve Fit, Import Parser Sections, Custom Calculation Functions, Generate Inventory Labels, Inventory Admin Permission
Read More