July 18, 2022
CDD Vault Update (July 2022): Mixture Lookups; Include Source and API Parameters to Limit the Amount of Data Returned
Vault Name is Included when Looking Up Mixture Components
When registering a new Mixture using the Vault Lookup interface, the source of the component is displayed to indicate where the component is currently registered.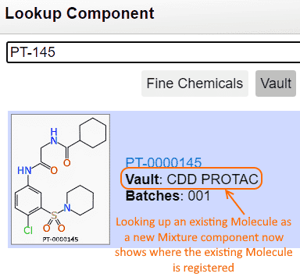
Limiting Data Returned Via API Calls
API parameters are available to limit the actual data returned by API calls. This can significantly improve the performance of these API endpoints. I. The "only_ids" parameter on the GET Molecules, GET Batches, GET ELN Entries and GET Protocols API calls allows you to return only the relevant object IDs instead of all associated data. This reduces the amount of data and time, and can be useful to determine if objects are deleted. TheGET Molecules API call, sending this JSON file:
{ "created_after":"2022-01-01",
"only_ids":"true"
}
Would return only these JSON results:
{
"count": 4,
"objects": [
109638855,
109641378,
109638813,
112228460
]
} II. The "no_structures" parameter on the
GET Molecules and
GET Batches API calls allows you to omit the structural information from the resulting JSON. This, too, allows for a faster response and avoids passing along sensitive structural information. The
GET Molecules API call, sending this JSON file:
{ "created_after":"2022-01-01",
"no_structures":"true"
}
Would omit all chemical structure information from the JSON results. III. The “molecule_fields” and “batch_fields'' parameters for the
GET Molecules and
GET Batches API let you specify exactly which fields you’d like to return. By default all fields are returned and you can specify a non-existent field, e.g. -1 to not return any fields. The
GET Molecules API call, sending this JSON file:
{ "names":"CAW-0000028",
"no_structures":"true",
"molecule_fields":"Source"
}
Would return only the data stored in the Source Molecule Field in the JSON results (data in other Molecule fields would be omitted):
"molecule_fields": { "Source": "Graceland CRO" },
Helpful Hint
Nearly all of the CDD API endpoints support a number of parameters which can be passed when making API calls. Historically, these parameters are passed by adding them directly to the url used for the API call. However, with the number of useful parameters growing, adding a combination of parameters to the url may make the url long and rather unwieldy. We'd like to recommend a "best-practice" method for passing parameters so they no longer have to be added to the url. Instead of adding parameters to the url, create a JSON file containing all of the parameters you wish to submit and then reference your JSON file in your API call.Other posts you might be interested in
View All Posts
CDD Blog
3 min
April 14, 2025
Let’s Talk Security - Why a Bug Bounty May Be More Valuable Than a Penetration Test
Read More
CDD Vault Updates
7 min
April 10, 2025
CDD Vault Update (April 2025): Biphasic Curve Fit, Import Parser Sections, Custom Calculation Functions, Generate Inventory Labels, Inventory Admin Permission
Read More
CDD Blog
9 min
April 8, 2025
Drug Discovery Industry Roundup with Barry Bunin — April 8th, 2025
Read More


