The Research Informatics and Development groups at CDD are continually developing computational tools within CDD Vault to help accelerate drug discovery. The newest addition to this suite of features is a novel Deep Learning model to aid medicinal chemists.
This model leverages nearest neighbor searches in combination with our deep learning derived chemically rich vectors (CRVs) to provide a computational model that accurately identifies similar public structures in a safe and secure environment. It gives you access to information about the chemical properties of molecules, their desired target properties/profiles, and enables you to assess commercial availability and/or retrosynthetic viability. 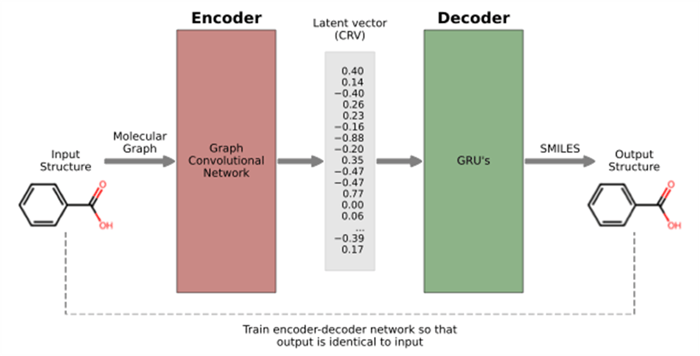
CDD deep learning architecture: Encoder (graph convolutional network) coupled with decoder (GRUs) through latent vector
Key aspects of the deep learning network architecture and its training are:
- The network is a combination of a graph convolutional network to represent a molecule as a chemically rich vector of length 384 and a generative model to derive structures matching a given CRV
- The network was trained using structures from ChEMBL version 28
- The network is trained using a combination of objectives, to ensure that the CRV represents structural information, covers the latent space well, and can be used to reconstruct the chemical structure represented by a CRV
- The CRV structure representation combined with a suitable distance metric, can be used for similarity searches that are complementary to established approaches
- Validation studies showed that the CRV can be used as an excellent descriptor in QSAR models. This coupled with the generative model optimizes properties by moving in the latent space.
In this release of our deep learning technology, we focus on the secure similarity search capability. For additional capabilities, please talk to us.
With the Deep Learning feature, users are able to discover compounds by rapidly searching ChEMBL for similar structures, all within CDD’s secure environment. CDD's proprietary Machine Learning suggests similar chemical structures which are biased toward commercially available, synthesizable molecules.
Once this feature is enabled in your CDD Vault, the "Find ChEMBL molecules using deep learning similarity" button is available from the Molecule Overview page.
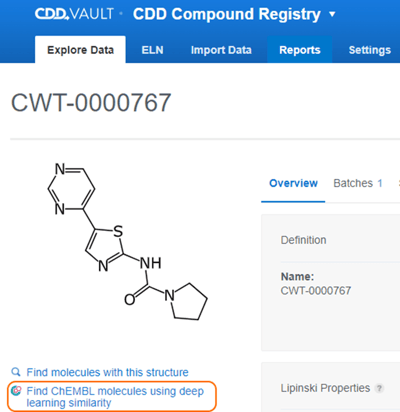
Clicking this button will present the user with a grid table view of similar Molecules from ChEMBL. There are also buttons to switch to a scaffold view and also export these Molecules.
Note: the export is a csv file containing the ChEMBL ID, SMILES string and SMILES string for the corresponding scaffold.
This blog is authored by members of the CDD Vault community. CDD Vault is a hosted drug discovery informatics platform that securely manages bothfivity-registration-system" rel="noopener" data-saferedirecturl="https://www.google.com/url?hl=en&q=https://info.collaborativedrug.com/cddchemreg&source=gmail&ust=1478041597053000&usg=AFQjCNEni2InjeGNLQZy3vgNFKTMZsYrmQ">chemical registration, data visualization, inventory, and electronic lab notebook capabilities.
Other posts you might be interested in
View All Posts
CDD Vault Updates
4 min
December 21, 2023
CDD Vault Update (December 2023 #3): New Password Algorithm and API Parameter Updates (ELN, Plates, Runs)
Read More
CDD Blog
4 min
April 8, 2021
CDD Awarded $1.5M Phase 2 SBIR Grant on an Intelligent Chemical Structure Browser for Drug Discovery and Optimization
Read More
CDD Vault Updates
2 min
February 26, 2024
CDD Vault Update (February 2024): New Models for LogD and LogP, Similarity Scores in Deep Learning, and Updated GET Users Call
Read More


